SAvEM3: Pretrained and Distilled Models for General-purpose 3D Neuron Reconstruction
We introduce SAEM2-SAvEM3 pipeline for general-purpose neuron reconstruction, which extends general-purpose auxiliary tasks for SAM and lifts into the 3D U-Net by full-stage distillation.
Hao Zhai*, Jinyue Guo*, Yanchao Zhang*, Jing Liu, Hua Han
Key Laboratory of Brain Cognition and Brain-inspired Intelligence Technology, Institute of Automation, Chinese Academy of Science
School of Future Technology, School of Artificial Intelligence, University of Chinese Academy of Sciences
Abstract
With an explosion in the uptake of volume electron microscopy (vEM) across neuroscience and fast-paced advances in imaging protocols, it is timely to introduce general-purpose automation for newly generated large-scale vEM datasets. Recent vision foundation models (e.g., SAM) set a new benchmark for the generalization of 2D segmentation. However, SAM has difficulty handling neurons that are densely packed into 3D volumes. To overcome this obstacle, we consider solutions from both data and model aspects. In terms of data, we introduce a data engine to optimize manual labeling, including (i) human-in-the-loop data cleansing and (ii) model-in-the-loop data unification. In terms of model, we present the SAEM\(^2\)-SAvEM\(^3\) with strategies, including (i) auxiliary learning, which predicts complementary representations for SAM masks and improves performance on dense instances; (ii) full-stage distillation, which integrates ViT embeddings into a 3D U-Net, achieves 2D-to-3D lifting and model slimming at the same time; and (iii) prompt-based graph-cut, which reuses SAM prompts to assign weights of nodes and edges in the oversegmentation graph. According to evaluations of dense and large-scale sparse groundtruth neurons, the out-of-distribution performance of our pretrained-distilled models is on par with the state-of-the-art supervised and semi-supervised methods. The overall pipeline provides a possible general-purpose solution for 3D neuron reconstruction in any new vEM data. Our code will be available at https://github.com/JackieZhai/SAvEM3.
Motivation
Vision foundation models have recently been well developed by introducing vision transformers (Segment Anything Model, SAM) and training on extremely large datasets (SA-1B). Conceptually, they achieved state-of-the-art out-of-distribution generalization because of two interconnected necessities: a powerful model with prompt engineering and a data engine with a number of high-quality and diverse masks. However, there are still obstacles in both the model and data aspects toward general-purpose 3D neuron reconstruction in vEM or scaling connectomics.
In data aspect, there is currently no comprehensive dataset collection and unification for vEM. Meanwhile, datasets from various imaging methods, animal species and brain regions are likely to have different manual labeling styles.
In model aspect, SAM still can not segment the wiring in the brain, such as the splits and merges of dense axons, dendrites, and the homogeneous (weak semantic) plasma membrane. Besides, Micro-SAM and Tri-SAM directly use IoU or confidence scores to propagate 2D masks across (cross-section) images. How to lift from 2D SAM to 3D is thus a problem to be solved.
Pipeline
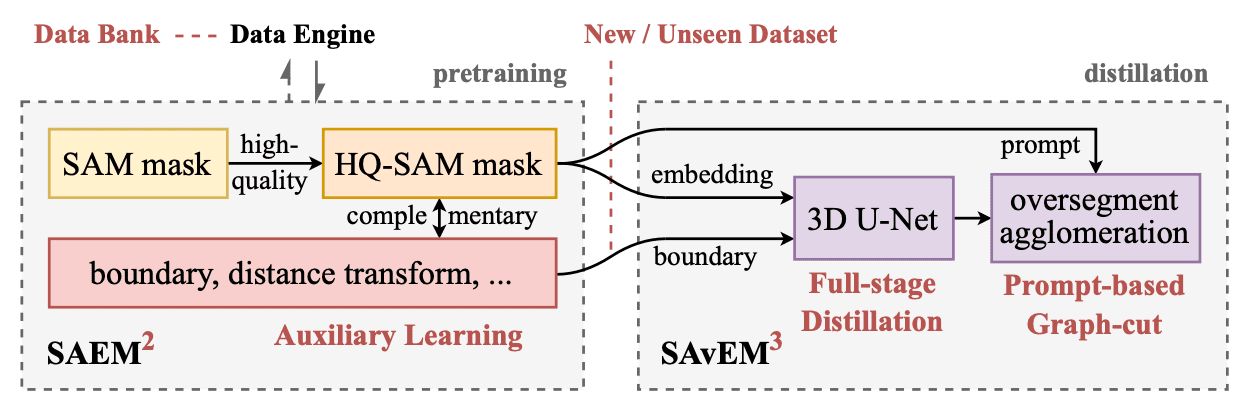 Overall pipeline of general neuron reconstruction for any vEM data.
Overall pipeline of general neuron reconstruction for any vEM data.
Our overall pipeline for general-purpose 3D neuron reconstruction is shown in Fig. 1. To sum up, there are two phases: SAEM\(^2\) and SAvEM\(^3\). In the first phase, we exploit the collected data bank to train a 2D pretrained promptable generalist model, and the data engine iteratively optimizes data and models. In the second phase, we employ the previously unseen data (without manual labeling) to distill a 3D U-Net specialist model for boundary maps and then do oversegmentation and agglomeration to get 3D instances. This pipeline is a possible bottom-up automated solution for any new vEM data.
SAEM$^2$
Data bank: diverse imaging methods and animal species
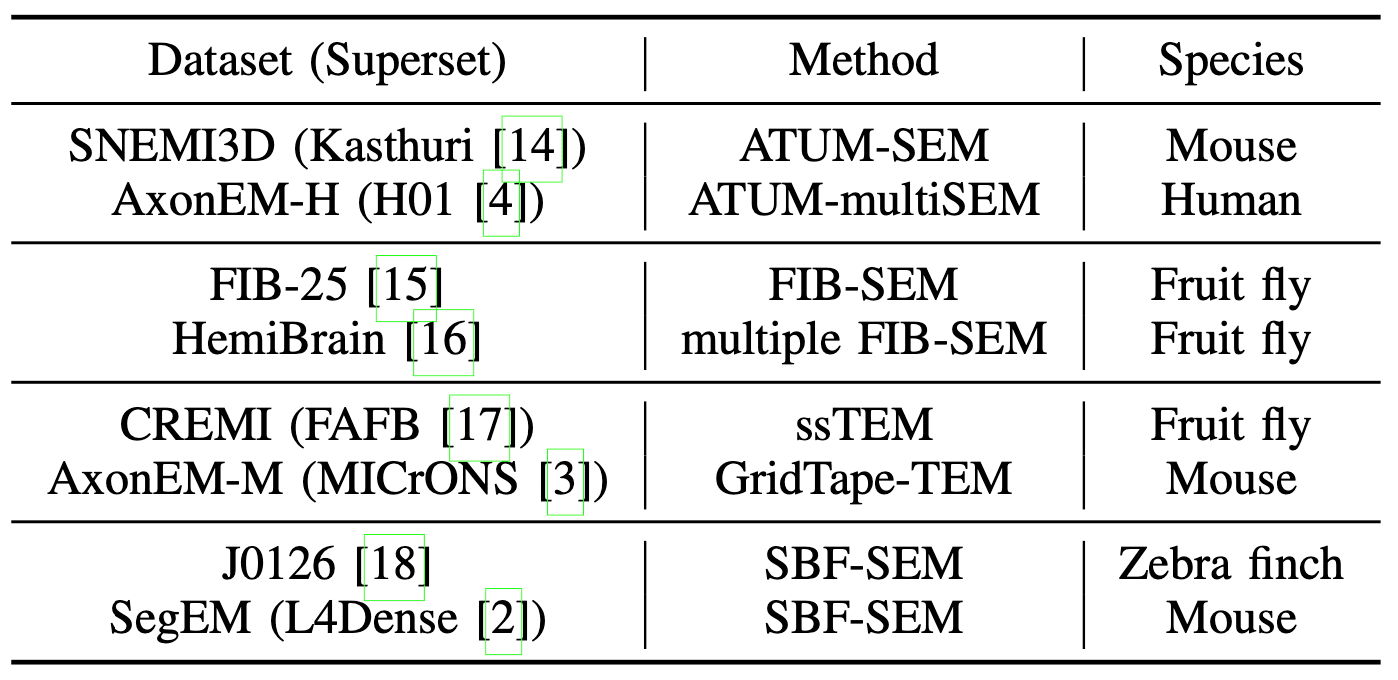 Diverse imaging methods and animal species in our data bank.
Diverse imaging methods and animal species in our data bank.
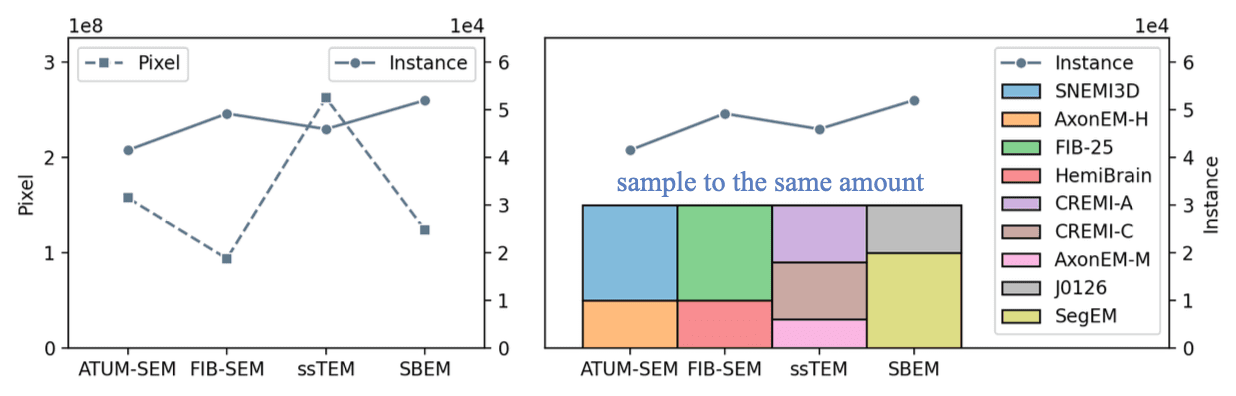 The SAEM\(^2\) training set picked from our data bank. Specified sampling numbers for each dataset accumulate to the same amount.
The SAEM\(^2\) training set picked from our data bank. Specified sampling numbers for each dataset accumulate to the same amount.
We have collected eight datasets from four different imaging methods and four animal species, and five of them are also open challenges. Note that all datasets have far larger supersets of raw data. Taking H01 as an example, roughly 0.001% of the data were labeled for training.
We then picked around 188K representative masks from the total of 649K masks to serve as the SAEM\(^2\) training set. We did this step because of the extremely fragmented fly neurites in FIB-SEM datasets and the unbalanced small volumes in SBEM datasets. Specifically, we chose 9 of 20 ATUM-SEM volumes, 2 of 3 FIB-SEM, 2 of 3 ssTEM, and 94 of 312 SBEM with nine volumes stitched into a volume. For each epoch during training, we randomly sample 30K instances from each imaging method to further balance them.
Data engine: unified labeling styles of the boundaries among four different imaging methods
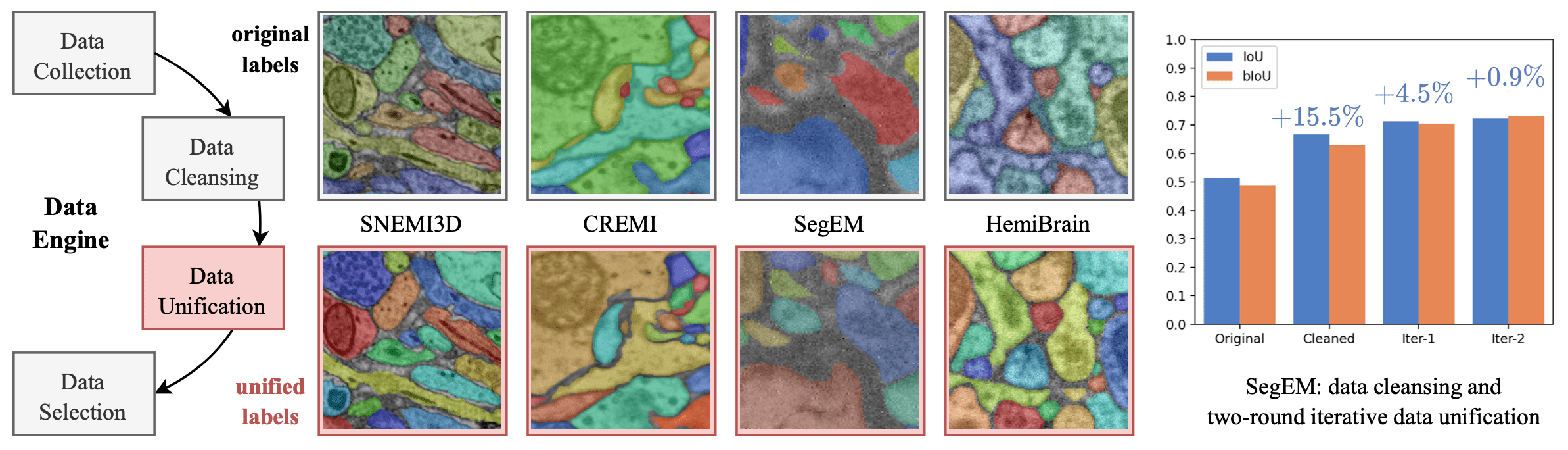 Data engine workflow. Middle: Examples of before/after unification labels from four different datasets. Right: The model performance improvement by data sampling, cleansing and two-round iterative unification. (Here, we use SegEM with the most diverse labeling style as a representative case.)
Data engine workflow. Middle: Examples of before/after unification labels from four different datasets. Right: The model performance improvement by data sampling, cleansing and two-round iterative unification. (Here, we use SegEM with the most diverse labeling style as a representative case.)
- Data cleansing (human-in-the-loop):
- adjust the xy-plane resolution of all datasets (including 4, 6, 8, 9 and 11.24nm) to 4nm for SAM and 8nm for HQ-SAM, SAEM\(^2\) and U-Nets.
- set the myelin of AC3/AC4 and AxonEM as backgrounds and masked the glia and defects of CREMI, FIB25, HemiBrain, and J0126.
- realigned the CREMI dataset and erode masks properly.
- check manually with an IoU threshold.
- Data unification (model-in-the-loop):
- use finetuned HQ-SAM and SAEM\(^2\) with NMS
- optimize the original boundary by prompted masks
- train the model iteratively with the new boundary
Auxiliary learning: hybrid representation of neural morphology for SAM
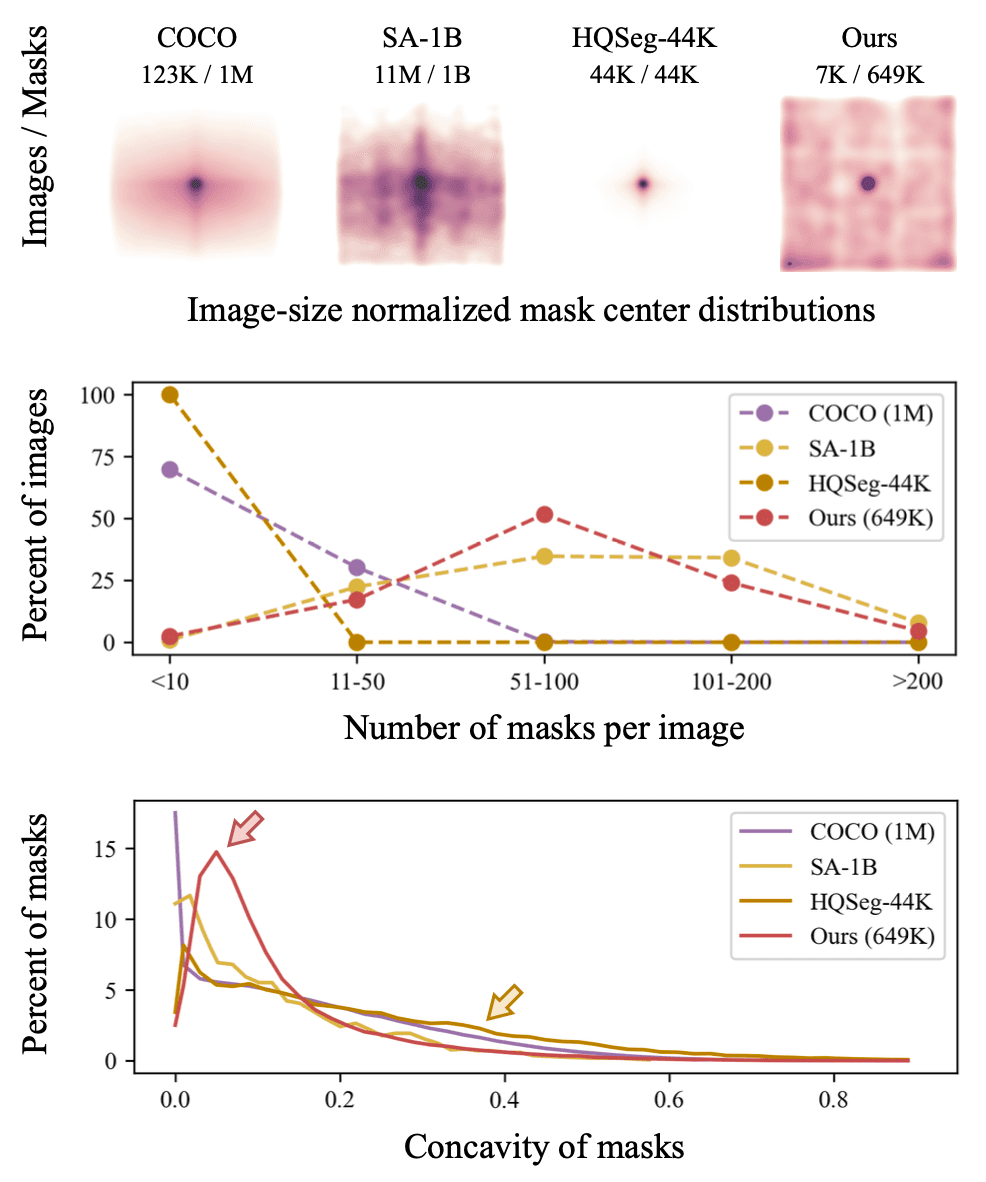 Properties of the COCO, SA-1B, HQSeg-44K and our datasets. SA is dense, HQSeg is concave, and neural morphology has characteristics of both.
Properties of the COCO, SA-1B, HQSeg-44K and our datasets. SA is dense, HQSeg is concave, and neural morphology has characteristics of both.
Background
The pretrained model SAM and HQ-SAM reflect the distribution of their training set, which are able to provide generalist model capabilities.
Compared to the classic dataset COCO,
- SA-1B: dense and uncentered,
- HQSeg-44K: thin and concave,
- Our Data Bank: dense, uncentered, thin, and even concave.
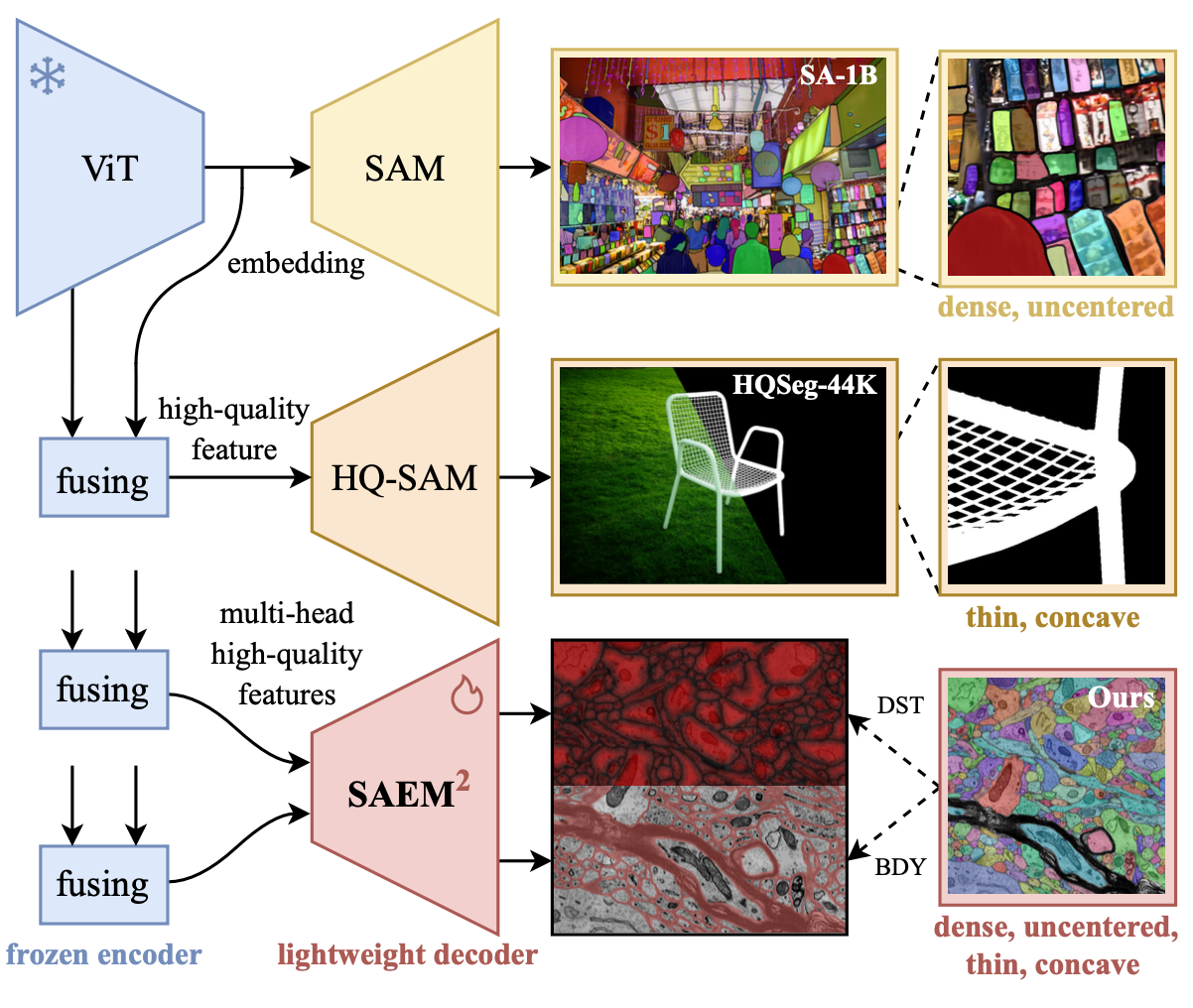 SAEM\(^2\) auxiliary learning. These images and masks indicate that the motivation for model design comes from the characteristics of data. (Examples from the SA-1B, HQSeg-44K and SNEMI3D dataset in our data bank.)
SAEM\(^2\) auxiliary learning. These images and masks indicate that the motivation for model design comes from the characteristics of data. (Examples from the SA-1B, HQSeg-44K and SNEMI3D dataset in our data bank.)
Improvement
To extract the hybrid representation of neural morphology, we present SAEM\(^2\) with auxiliary learning tasks including the plasma membrane as boundaries (BDY) or distance transformation (DST).
It is worth mentioning that another side to the story of SAEM\(^2\) contributes to the adaption to widely-used bottom-up methods of 3D neuron reconstruction. The bottom-up methods include steps of binary pixel-wise classification (e.g., boundaries), watershed-based oversegmentation and the agglomeration to instance segmentation. In this case, our auxiliary decoders provide the results of pixel-wise classification (i.e., BDY or DST) for next-step oversegmentation, which are not available in the original SAM or HQ-SAM.
SAvEM$^3$
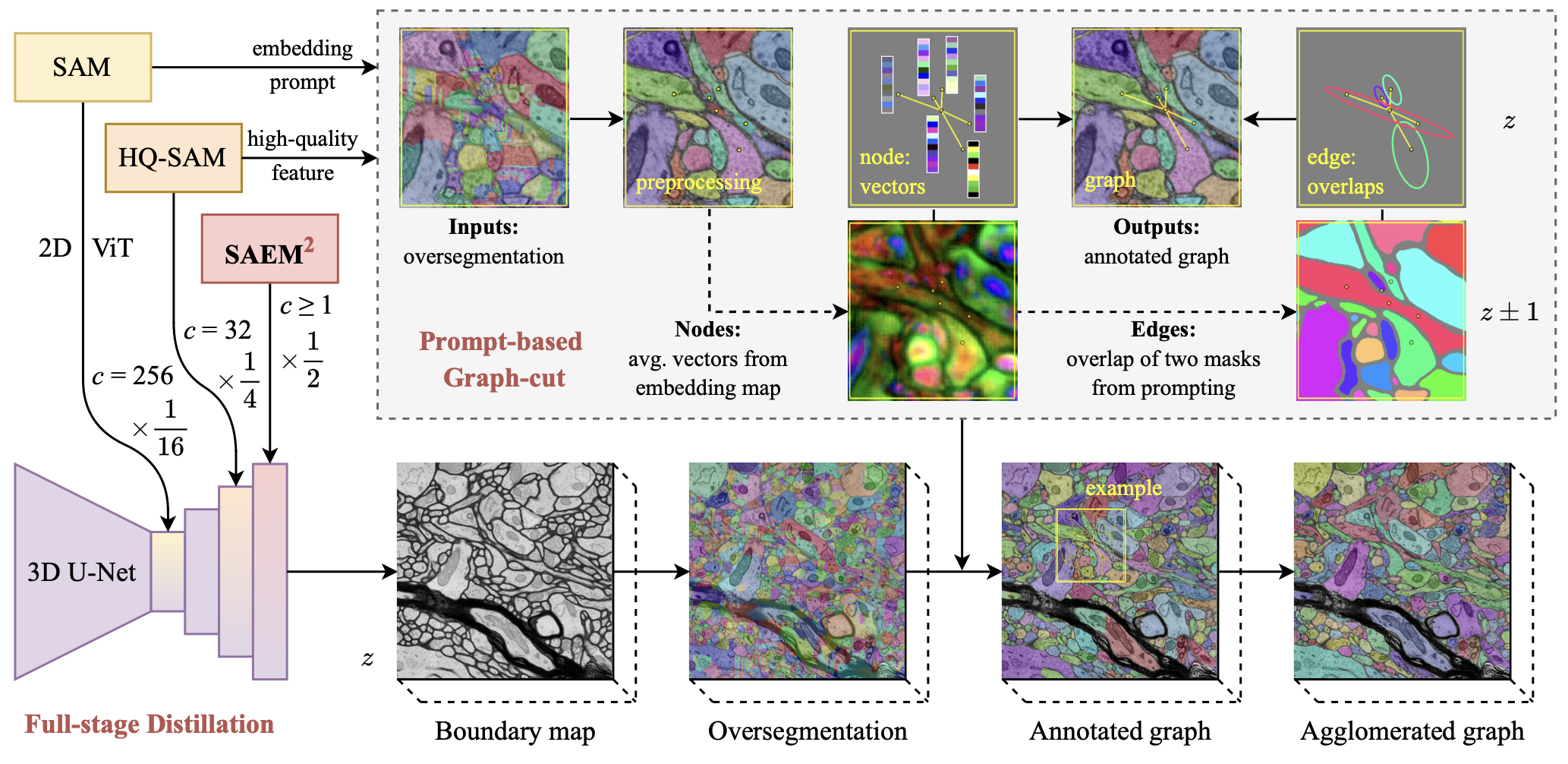 SAvEM\(^3\) full-stage distillation and prompt-based graph-cut. Left: The 3D U-Net is simultaneously distilled from global SAM embeddings, local HQ-SAM features and SAEM\(^2\) boundaries. (\(c\) is the number of channels. 1/16, 1/4 and 1/2 are x and y-axis downsample ratios.) Top-right box: The graph-cut reuses HQ-SAM features and SAM prompting as extra nodes and edges. (z±1 means that the prompting seeds are also from two adjacent sections.)
SAvEM\(^3\) full-stage distillation and prompt-based graph-cut. Left: The 3D U-Net is simultaneously distilled from global SAM embeddings, local HQ-SAM features and SAEM\(^2\) boundaries. (\(c\) is the number of channels. 1/16, 1/4 and 1/2 are x and y-axis downsample ratios.) Top-right box: The graph-cut reuses HQ-SAM features and SAM prompting as extra nodes and edges. (z±1 means that the prompting seeds are also from two adjacent sections.)
We propose SAvEM\(^3\) with two strategies: (1) full-stage distillation, integrates ViT embeddings and SAEM\(^2\) predictions into a 3D U-Net, achieves 2D-to-3D lifting and model slimming at the same time (left); (2) prompt-based graph-cut, reuses SAM prompting to assign weights of nodes and edges for the agglomerated graph (top-right box).
Specifically, we implement the state-of-the-art “Superhuman” 3D Residual U-Nets to serve as the student network. For a brief comparison of model parameters, the SAM (ViT-H, serves as the teacher network for SAvEM\(^3\) distillation) has about 615M, the distilled Mobile-SAM (Tiny-ViT) has about 9M, yet the 3D Res U-Net only has about 7M parameters.
Results
Comparison with General-purpose Methods
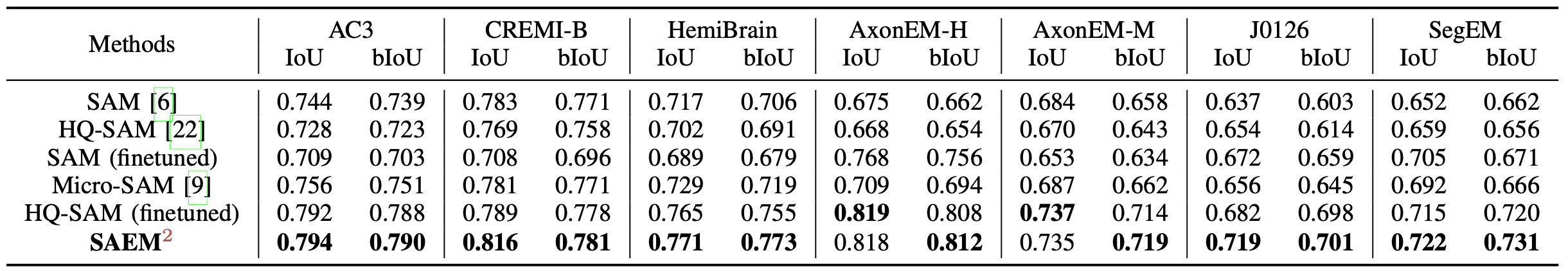 Evaluation of SAEM\(^2\) and other original/finetuned SAM variants by IoU \(\uparrow\) and bIoU \(\uparrow\) metrics.
Evaluation of SAEM\(^2\) and other original/finetuned SAM variants by IoU \(\uparrow\) and bIoU \(\uparrow\) metrics.
Comparison with State-of-the-art Methods
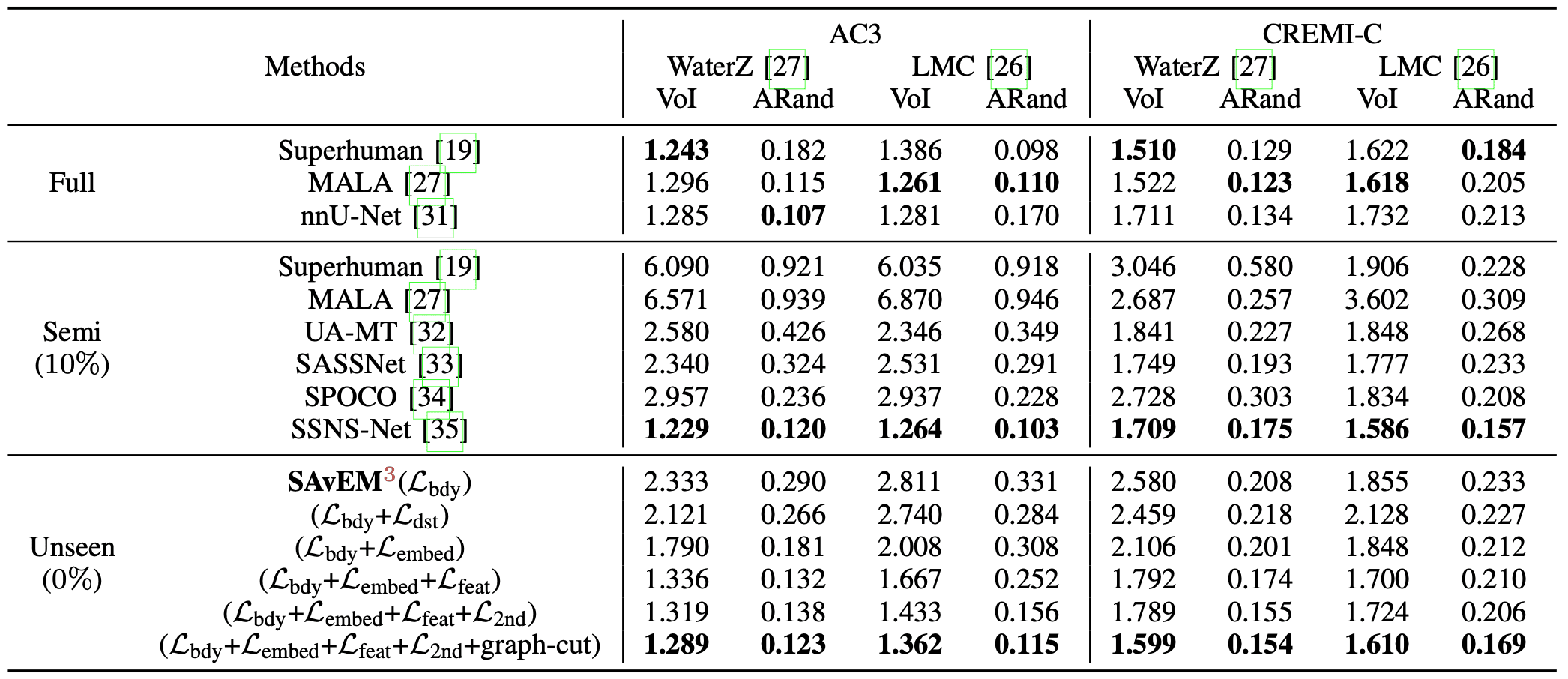 Evaluation of SAvEM\(^3\) and other state-of-the-art supervised/semi-supervised methods by VoI \(\downarrow\) and ARand \(\downarrow\) metrics.
Evaluation of SAvEM\(^3\) and other state-of-the-art supervised/semi-supervised methods by VoI \(\downarrow\) and ARand \(\downarrow\) metrics.
Ablation Studies
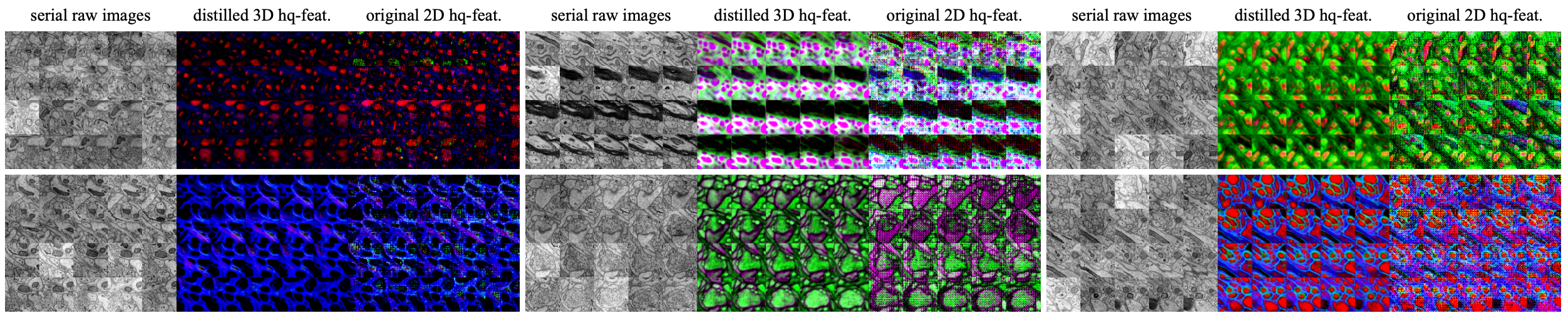 Visualization of SAvEM$^3$$ full-stage distillation. Here, we present examples of high-quality feature 2D-to-3D lifting with smoothing regularization.
Visualization of SAvEM$^3$$ full-stage distillation. Here, we present examples of high-quality feature 2D-to-3D lifting with smoothing regularization.
Citation
Zhai H, Guo J, Zhang Y, Liu J, Han H (2024). SAvEM3: Pretrained and Distilled Models for General-purpose 3D Neuron Reconstruction. In Proceedings of IEEE International Conference on Bioinformatics and Biomedicine (BIBM). accepted.