SegNeuron: 3D Neuron Instance Segmentation in Any EM Volume with a Generalist Model
We introduce SegNeuron, a generalist model for dense neuron instance segmentation with customized strategies, including pretraining via multi-scale Gaussian mask reconstruction, domain-mixing finetuning, and foreground-restricted instance segmentation.
Yanchao Zhang*, Jinyue Guo*, Hao Zhai*, Jing Liu, Hua Han
Key Laboratory of Brain Cognition and Brain-inspired Intelligence Technology, Institute of Automation, Chinese Academy of Science
School of Future Technology, School of Artificial Intelligence, University of Chinese Academy of Sciences
Abstract
Building a generalist model for neuron instance segmentation from electron microscopy (EM) volumes holds great potential to accelerate data processing and analysis in connectomics. However, the diversity in visual appearances and voxel resolutions present obstacles to model development. Meanwhile, prompt-based foundation models for segmentation struggle to achieve satisfactory performance due to the inherent complexity and volumetric continuity of neuronal structures. To address this, this paper introduces SegNeuron, a generalist model for dense neuron instance segmentation with strong zero-shot generalizability. To this end, we first construct a multi-resolution, multi-modality, and multi-species volume EM database, named EMNeuron, consisting of over 22 billion voxels, with over 3 billion densely labeled. On this basis, we devise a novel workflow to build the model with customized strategies, including pretraining via multi-scale Gaussian mask reconstruction, domain-mixing finetuning, and foreground-restricted instance segmentation. Experimental results on unseen datasets indicate that SegNeuron not only significantly surpasses existing generalist models, but also achieves competitive or even superior results with specialist models. Datasets, codes, and models are available at https://github.com/yanchaoz/SegNeuron.
Motivation
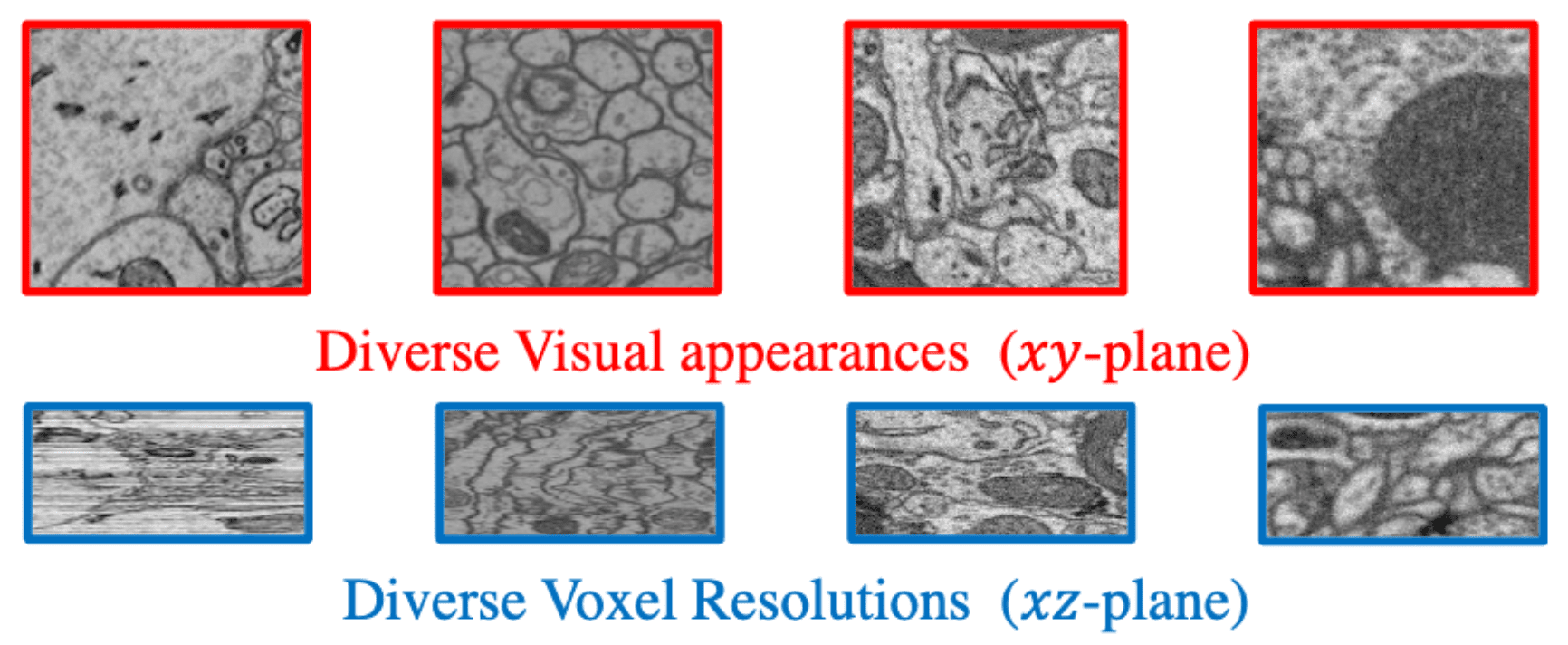 Challenges in generalist model development. Note that xz-plane here not scaled to isotropic.
Challenges in generalist model development. Note that xz-plane here not scaled to isotropic.
Recently, visual foundation models via prompt engineering have gained significant attention for segmentation tasks of biomedical images (e.g., MedSAM, MicroSAM). While they appear to integrate seamlessly into instance-based pipelines, the complexity of neural structures and the homogenization of EM images pose challenges in identifying neuron instances. Importantly, these 2D models struggle with the extensive splitting and merging of neurons in 3D space.
Fortunately, neuronal membranes demonstrate consistent characteristics across datasets, enabling the development of a generalist model to predict boundary descriptors (i.e., affinity maps) for 3D neuron reconstruction. We introduce SegNeuron, a boundary-based neuron segmentation model generalized across diverse data distributions and spatial resolutions. Such strong adaptability hinges on a diversified database and customized training strategies.
Datasets
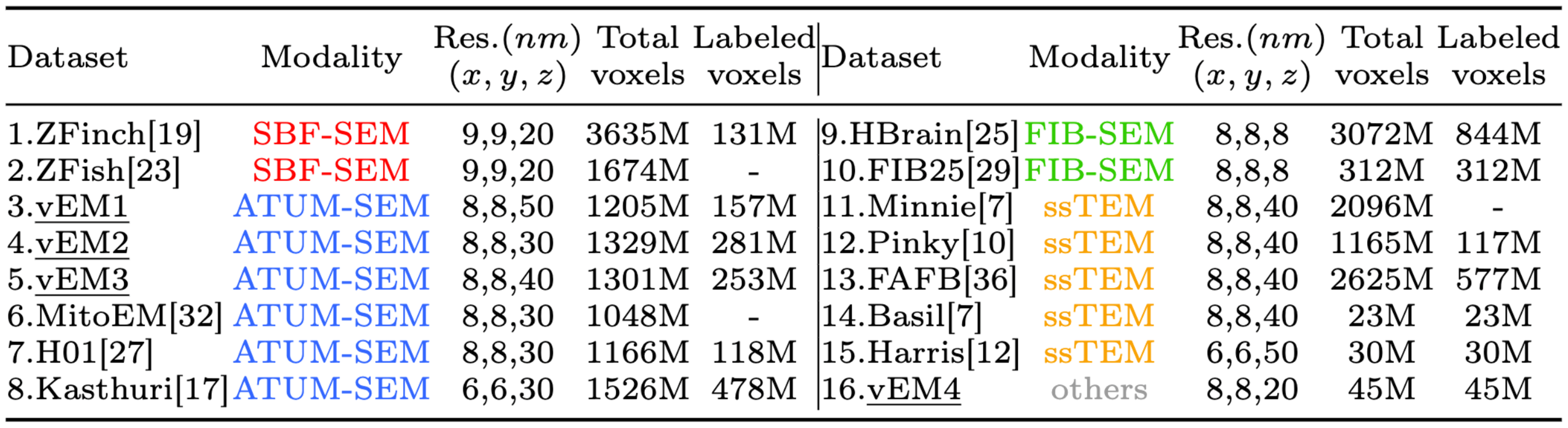 Details of our datasets, EMNeuron. Underlined items represent in-house datasets.
Details of our datasets, EMNeuron. Underlined items represent in-house datasets.
We integrate 16 volume EM datasets (4 in-house, 12 publicly available), covering diverse species, sample preparation protocols, imaging techniques, and voxel resolutions.
Datasets 1-13 are used for model development, and 14-16 for evaluation. We conduct comprehensive data cleaning and transforming, which includes adjusting tangential resolutions \((x,y)\) to 6-9 nm, unifying membrane thickness, and masking out unlabeled myelin and glial regions.
Notably, the overall size of some datasets reaches the petabyte level, making it impractical to utilize all for training. Therefore, we first select representative and informative unlabeled areas from each dataset and integrate them with the labeled parts to construct our dataset.
Methods
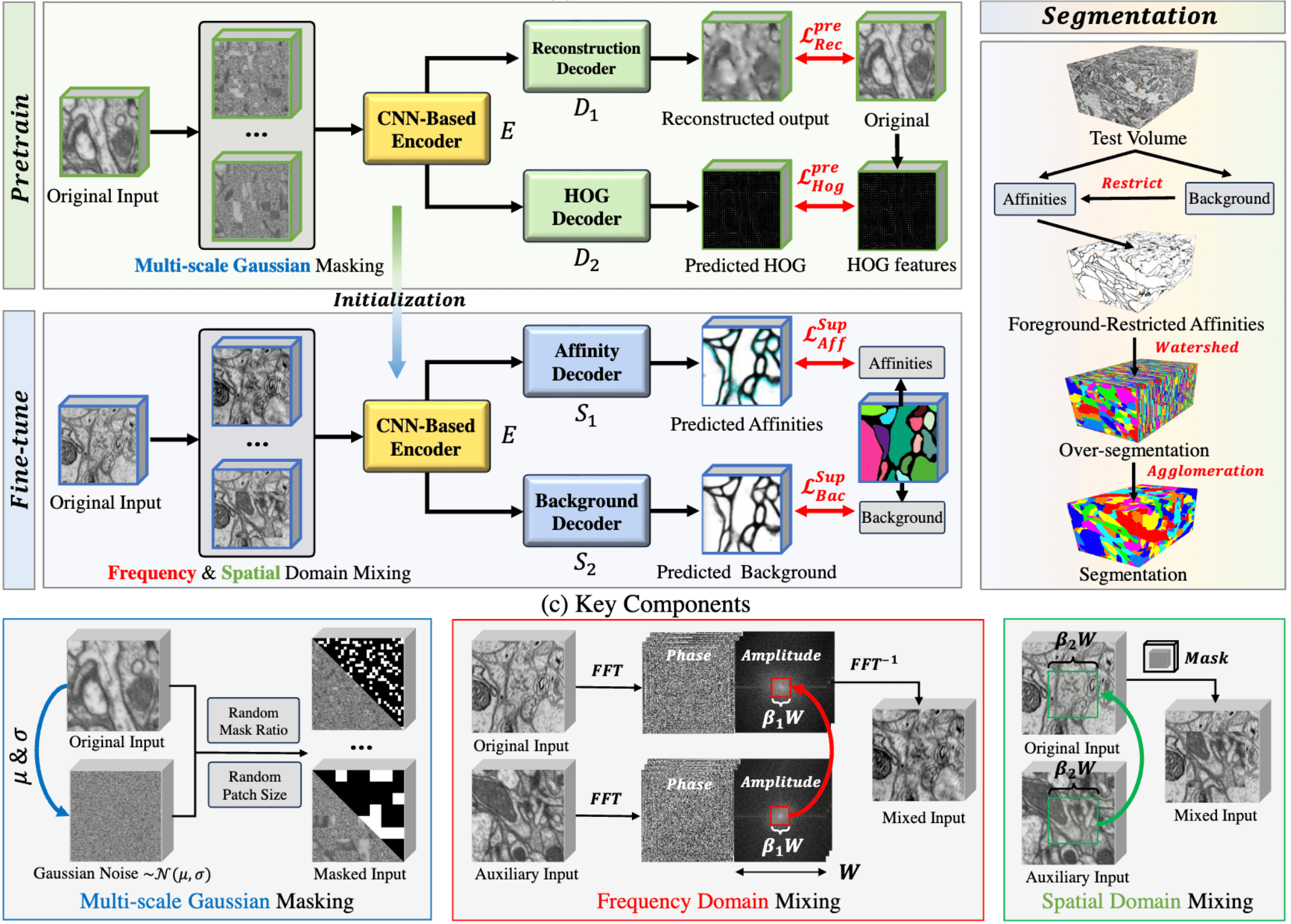 Customized pretraining and finetuning strategies of the main workflow for SegNeuron.
Customized pretraining and finetuning strategies of the main workflow for SegNeuron.
Pretraining: masking voxels with random Gaussian noise in a multi-scale manner
The masked input is then fed into the network consisting of an encoder \(E\) and two decoders \(D_1\). \(D_1\) is utilized to reconstruct the original input. For \(D_2\), we adopt the histogram of oriented gradients (HOG) as an additional prediction target, which proved to be effective in extracting high-frequency features. We used the mean squared error (MSE) loss for \(\mathcal{L}^{pre}\).
Finetuning: mixing volumes in both frequency and spatial domains
We initialize the encoder \(E\) in the segmentation network with pretrained weights. Subsequently, the entire model is finetuned in a supervised manner.
To encourage learning of generalized knowledge for neuron affinities and avoid overfitting to dataset-specific biases, we introduce two customized mixing strategies in the frequency and spatial domains, respectively.
In the frequency domain, the amplitude spectrum represents low-level features such as texture and appearance, while the phase spectrum captures higher-level content such as shape and boundary. Hence, for each layer of the 3D input, we preserve the phase spectrum and replace the low-frequency amplitude component with others. This operation preserves discriminative boundary information while significantly enriching the style and texture of the training data.
In the spatial domain, we extend CutMix (see SL-SSNS) to the neuron segmentation task, generating new inputs with mixed semantic contents.
During training, we predict both neuron affinities and foreground masks via two decoders \(S_1\) and \(S_2\)We used the cross entropy loss for \(\mathcal{L}^{sup}\).
Instance Segmentation: foreground-restricted segmentation
To mask noise values, we filter background voxels of predicted affinity maps using predicted foreground masks as the threshold. Such foreground-restricted affinities then serve as input for graph-based instance segmentation.
Results
- In-distribution: FIB25, Kasthuri (SNEMI3D), FAFB (CREMI)
- Out-of-distribution: Harris, Basil, vEM4
We use the variation of information (VI) and adapted random error (ARE) to evaluate instance segmentation (both lower values better).
Network Architectures and Pretraining Schemes
 Qualitative results of different network architectures and pretraining schemes.
Qualitative results of different network architectures and pretraining schemes.
Comparison with Generalist Models
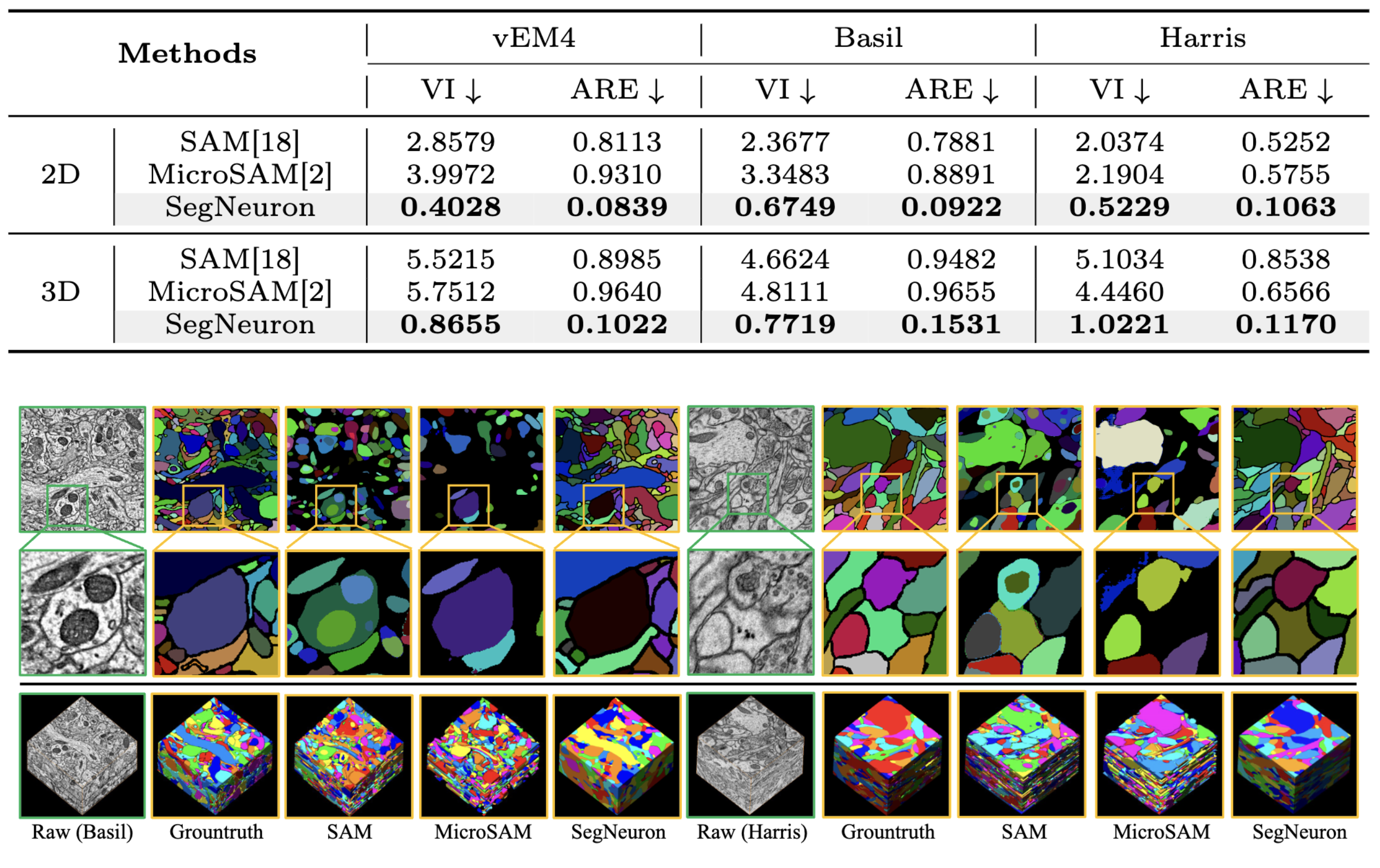 Quantitative comparison of the generalist models. Images are 2D and 3D visual comparison of the generalist models.
Quantitative comparison of the generalist models. Images are 2D and 3D visual comparison of the generalist models.
Comparison with Specialist Models
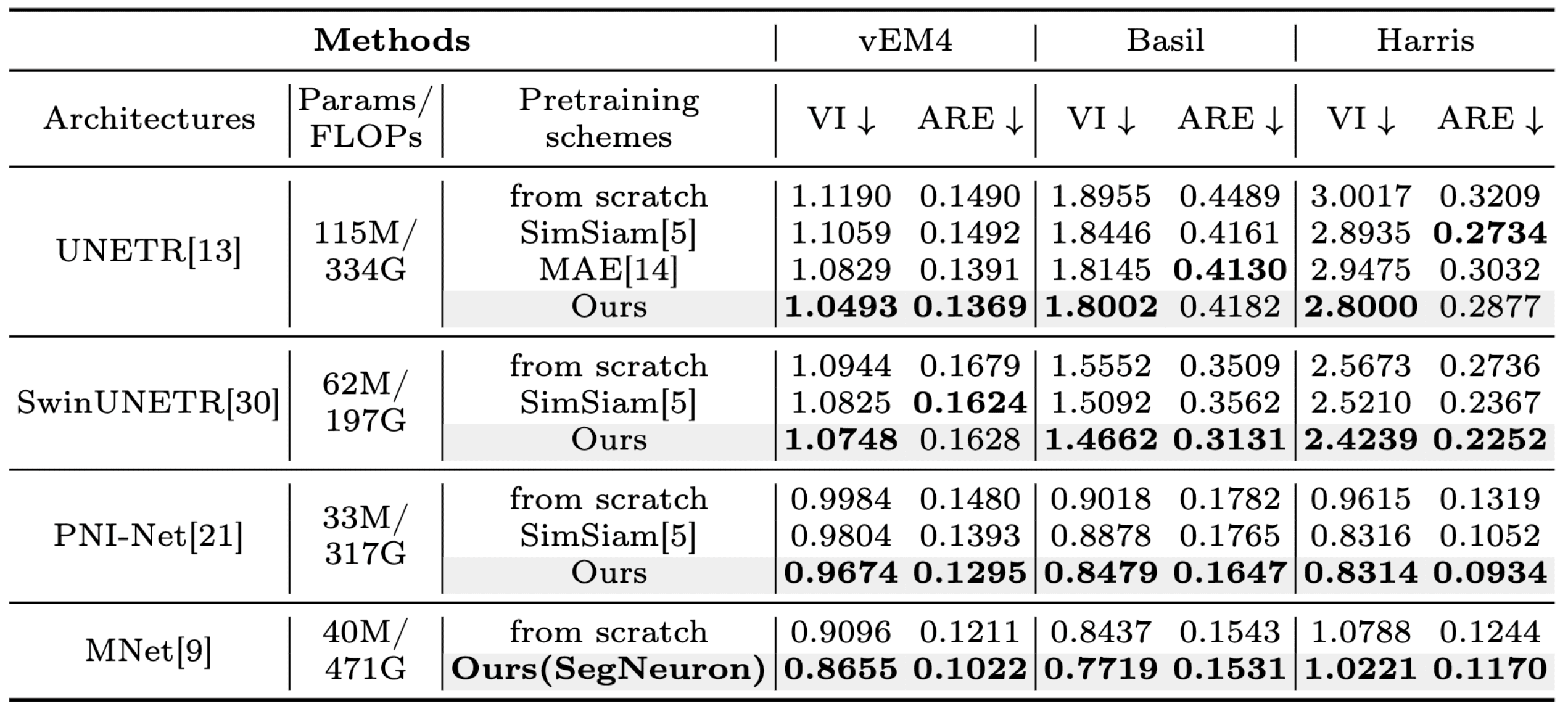
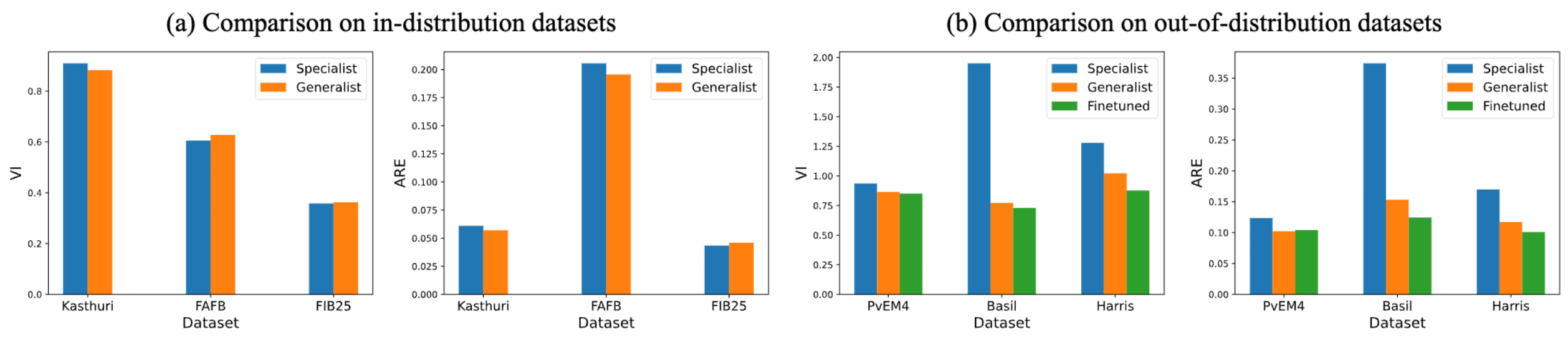 Quantitative comparison with specialist models.
Quantitative comparison with specialist models.
Ablations
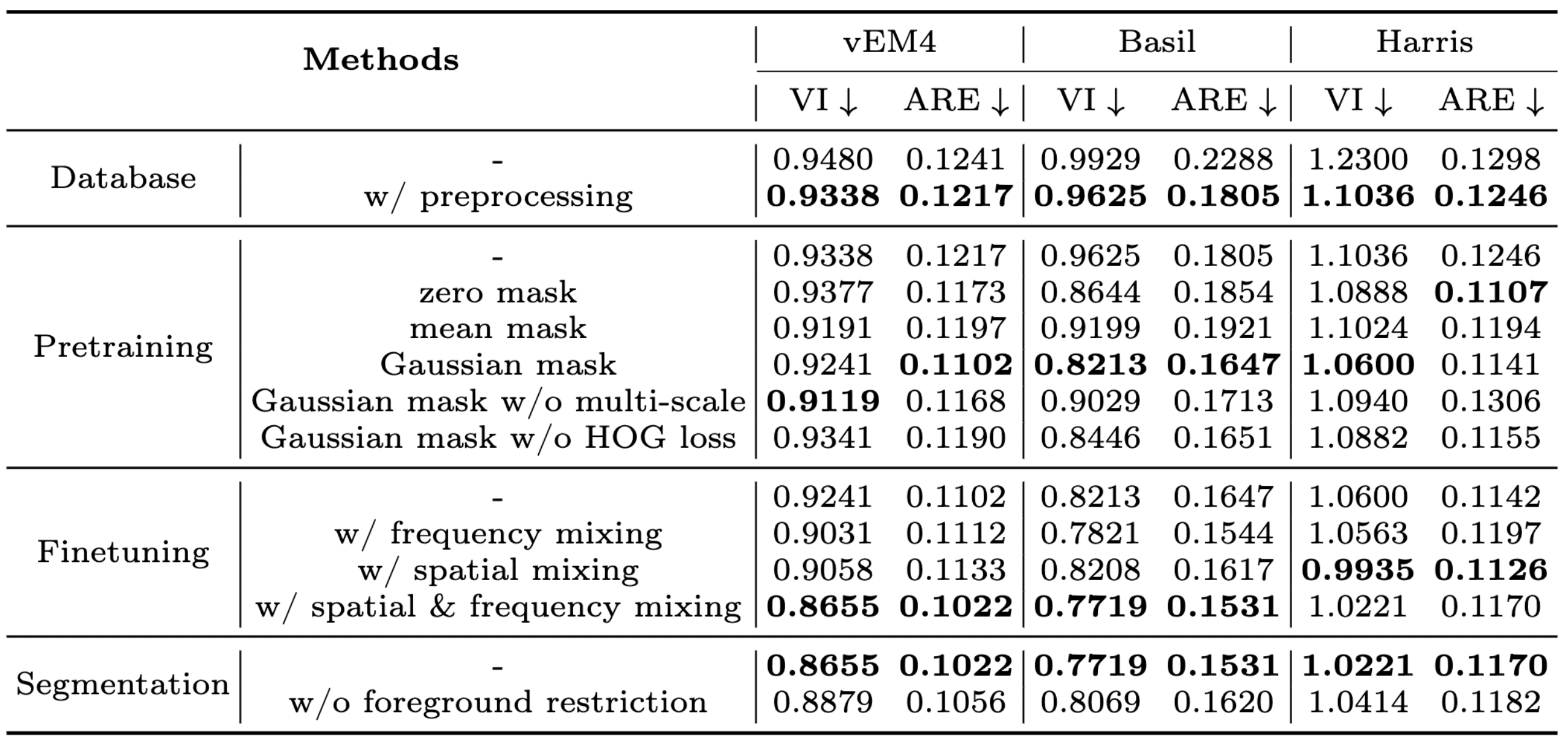 Ablation study for the generalist model development.
Ablation study for the generalist model development.
- Our pretraining strategy effectively leverages multi-scale perception and reduces statistics distortion, making it well-suited for neuron segmentation.
- The mixing strategies in spatial and frequency domains encourage domain-invariant learning and enhance model generalization.
- Preprocessing and foreground-restricted segmentation significantly improve performance without extra overhead.
Citation
Zhang Y, Guo J, Zhai H, Liu J, Han H (2024). SegNeuron: 3D Neuron Instance Segmentation in Any EM Volume with a Generalist Model. In Proceedings of Medical Image Computing and Computer Assisted Intervention (MICCAI), 589-600. https://doi.org/10.1007/978-3-031-72111-3_55.